What do we mean by “Target Organism” in Target Malaria’s gene drive research?

I’m delighted to share a new paper that was published last week in Trends in Biotechnology, co-authored by Target Malaria team members, external experts in genetically modified organism (GMO) risk assessment and myself. The paper explores the implications of using gene drive in a ‘species complex’ such as An. gambiae s.l. Because species within the complex are closely related genetically, they can still successfully mate with each other to produce fertile offspring or ‘hybrids’, albeit at low frequencies. This means that release of gene drive in one species of the complex could lead to transfer of that gene drive to other sibling species via hybridisation. That creates choices about what we might consider to be the ‘target organism’ of our gene drive.
Target Malaria is researching the use of the gene drive technology as a tool to modify mosquitoes to reduce the transmission of malaria by targeting An. gambiae s.l., a complex of at least nine species; six of which are known to be substantial transmitters, or vectors, of malaria in Africa.
Gene drives share many environmental risk assessment (ERA) considerations as other genetic modifications but they can differ significantly in their potential to spread, increase in frequency, and persist in target populations. Unlike other genetic modifications, gene drives have the ability to cause a selected trait to spread rapidly through a species.

In the wild and in laboratory settings, sibling mosquito species can successfully mate to produce viable offspring, regardless of whether they are vectors or not. Importantly, females, but not males, of these offspring can be fertile. Nonetheless, the likelihood of finding such hybrid mosquitoes in field samples varies greatly between different combinations of species.
According to some field studies, typically, only about 0.1% of mosquito collected in the wild could be An. gambiae s.s./An. coluzzii hybrids. In addition, some species that do not overlap geographically, and therefore would not come into direct contact, cannot produce hybrids in the field. This includes An. melas, which is found along the coast of West Africa, and An. bwambae, which is restricted to hot springs in the Toro District of Uganda.
This means that the gene drive could eventually transfer to all sibling species of the complex, both by direct hybridisation between geographically-overlapping species and, indirectly, by transferring from one species to another overlapping ones like stepping-stones until the gene drive was transferred to all species of the complex, including to the likes of An. melas and An. bwambae.
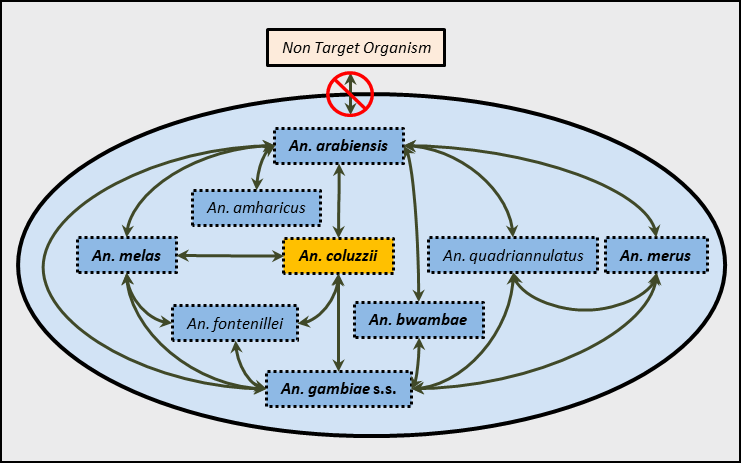
So all of this leads to the following questions:
- What species would we targeting with our gene drive?
- From a mechanistic point of view, would we say the gene drive would target all members of the An. gambiae s.l. species complex, whether vectors or non-vectors?
- From an intentional point of view, what species would we be targeting with the gene drive? Only vectors? Or non-vectors too?
- Or given some of the uncertainties of transfer of gene drive to other sibling species, would we be targeting just the species that we have released that contain the gene drive?
- In other words, what is the Target Organism?
This is an important concept to resolve for any ERA, including for our gene drives. Once we define what we mean by Target Organism, this would then establish what we mean by Non-Target Organisms, which would be species that are not direct targets of the gene drive. These definitions would form the basis of how we could evaluate potential efficacy of the gene drive versus potential harm from the gene drive.
Using our doublesex-based gene drive as a specific case study, we worked with a number of GMO risk assessment experts over the last year to develop four options on defining Target Organisms for gene drives in species complexes.
One of these options involves the proposal of the new concept of ‘Target Species Complex (TSC)’ in order to accommodate the unusual ‘porous’ species boundaries found in the An. gambiae s.l species complex that all for hybridisation.
Here, the TSC itself could be considered as the ‘functional biological entity’ that is the vector of disease transmission – not individual species – and thus this TSC could simultaneously be considered as both the intentional and mechanistic target of the gene drive.
Applying such nuanced thinking to the assessment of gene drive in species complexes should strengthen our understanding of any potential risks from the potential use of gene drive in the field, and itself represents a step forward on the path to development of novel malaria vector control tools.
